-
PRABI-GERLAND RHONE-ALPES BIOINFORMATIC POLE GERLAND SITE -
Institute of Biology and Protein Chemistry -
 A brief introduction to SOPM
A brief introduction to SOPM
SOPM (Self-Optimized Prediction Method) is a secondary structure prediction method based on the homologue method described by
Levin et al.. A new set of parameters have been optimized on a non-redundant databank of 510 sequences derived from Hobohm and
Sander (PDB select 25% August 1999).
The computing time has been divided by 3.
NPS@ is the original server for this method.
 Availability in NPS@
Availability in NPS@
This method is available :
 Parameters
Parameters
You can set the number of conformational states to predict : 3 or 4.
The similarity threshold parameter is the threshold below which a subject peptide is rejected when it's compared with a query
peptide of the sequence.
The window size parameter sets the length of the peptides to use.
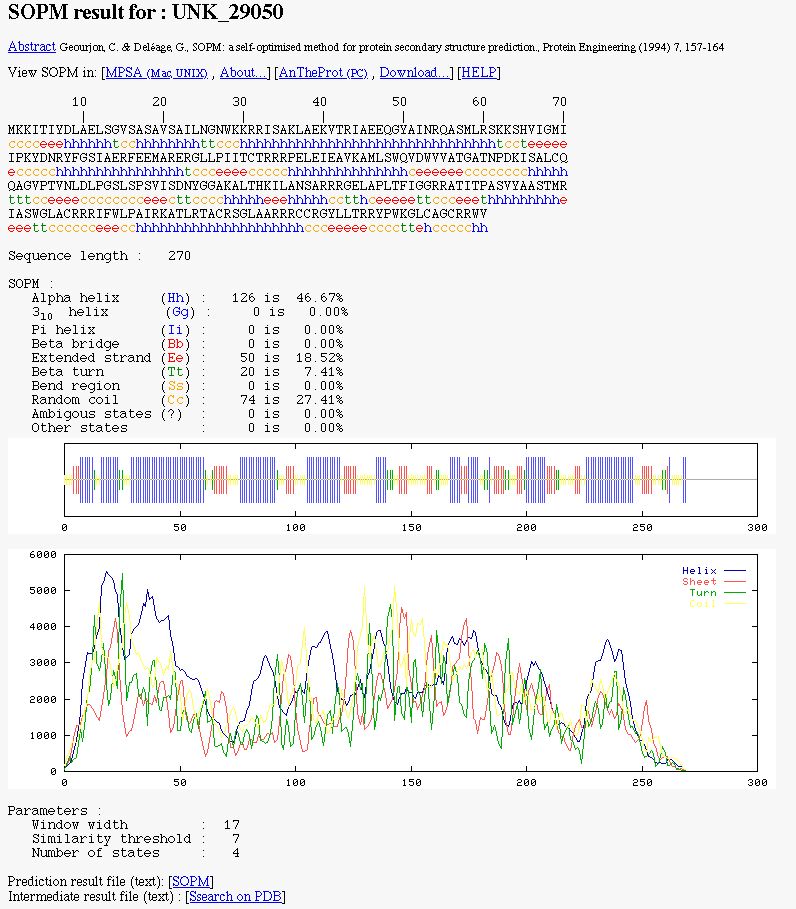
 NPS@ SOPM output example
NPS@ SOPM output example
You can see: