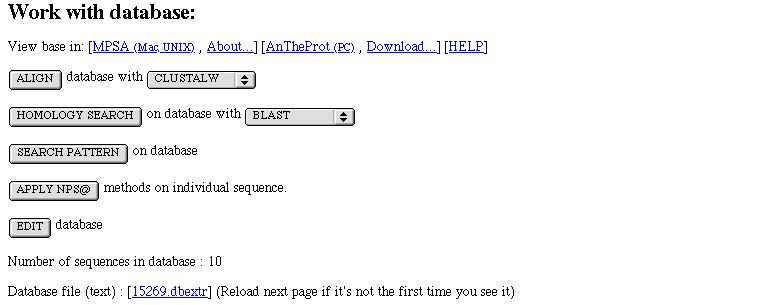
Work with a database
 In NPS@ you can work with a database.
In NPS@ you can work with a database.
The database can have four origins :
Only protein databases in Pearson/FASTA format are currently
supported by NPS@.
 What can you do then ?
What can you do then ?
You can :
- View it it in MPSA/
ANTHEPROT
- Align it with CLUTALW or
MULTALIN if there are no more than 250,000 characters in the database.
- Do a homology search. The database will be the query one.
- Search the database for one or several pattern (
PATTINPROT).
- Apply NPS@ methods on each sequence of the database. You will then work with
individual sequences.
- Edit the database. It allows you to select/unselect sequence of the database. It writes a new database.
- Know the number of sequence in the database.
- See the database text file.
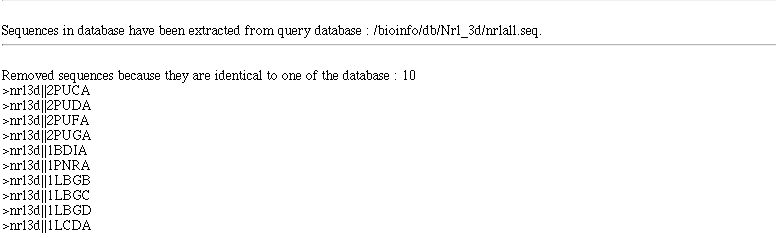
 When you come from an extract (After BLAST,PATTINPROT,...) few more informations are displayed.
When you come from an extract (After BLAST,PATTINPROT,...) few more informations are displayed.
- The database on which full sequence extraction has been made or it says you that it come from a partial extraction.
- Sequences identifier of the sequence removed in built database (when this option has been selected).
 In NPS@ you can work with a database.
In NPS@ you can work with a database. In NPS@ you can work with a database.
In NPS@ you can work with a database. What can you do then ?
What can you do then ?
 When you come from an extract (After BLAST,PATTINPROT,...) few more informations are displayed.
When you come from an extract (After BLAST,PATTINPROT,...) few more informations are displayed.