A brief introduction to PHD
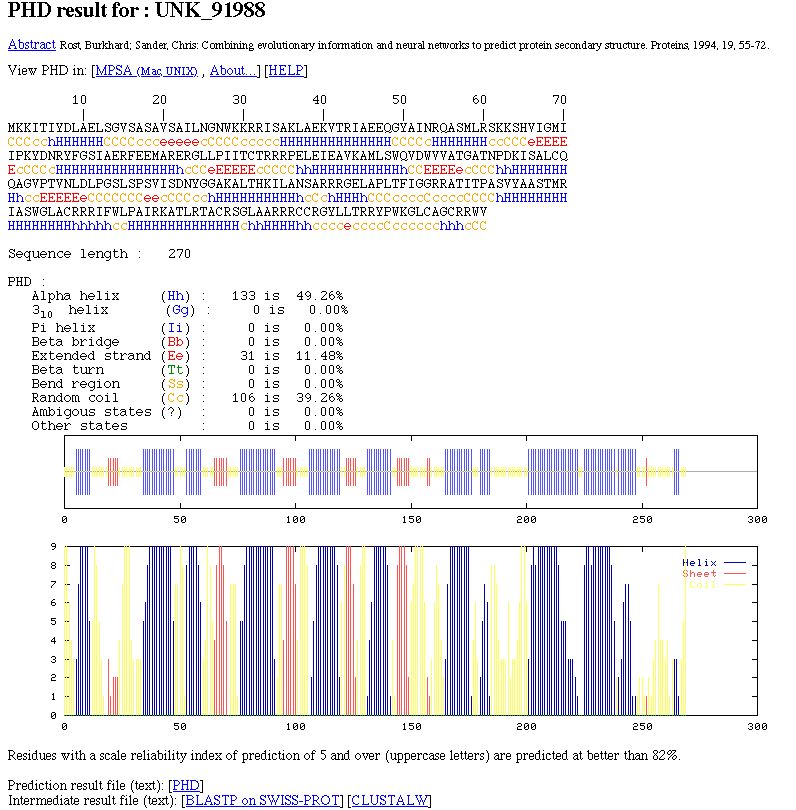
A brief introduction to PHDPHD are neural network systems (a sequence-to-structure level and a structure-structure level)to predict secondary structure (PHDsec), relative solvent accessibility (PHDacc) and transmembrane helices (PHDhtm). We only use PHDsec here. PHDsec focuses on predicting hydrogen bonds. The use of the evolutionary information held by a multiple sequence alignment increases the prediction accuracy. In the PredictProtein server, all is done with BLASTP and MaxHom. MaxHom filters the BLASTP result with a profile-based multiple alignment method. In NPS@, we run BLASTP, filter it and then run CLUSTALW. The multiple alignment is the input of the neural network. The PHD prediction done with NPS@ is better than the PHD prediction on the single sequence. But it's not exactly the same and could be a little bit less accurate than the PredictProtein one.
Warning : It can take up to 4 minutes to compute PHD for a sequence (22 seconds for RBTR_KLEAE (270 aa) and 3'25 minutes for MDR3_HUMAN (1270 aa) ). So, be careful when using it in alignment (the total computing time can't be above 3 hours on NPS@).
ACKNOWLEDGMENTS:
We thank Burkhard Rost for providing us PHD program and helping us to install PHD in NPS@ server.